Homology Modelling of Protein Structures
Homology Modelling of Protein Structures (step by step guide - Replico)
There are more than 15 million non-redundant protein sequences in the UniProt database but so far the number of structures is only about 180207.
So computational structure prediction is a very handy way of getting fairly good structures to get some valuable information about the protein.
Homology Modelling is a comparative structure prediction method.
What is Homology Modelling?
Homology modelling is basically mapping the structures of an unknown sequence based on the structure of a known sequence.
It is based on the principle that if two proteins share a high enough sequence similarity, they are likely to have very similar three-dimensional structures.
These structures then further help in :
- Understanding substrate and ligand binding in the case of enzymes.
- can be used in biochemical protein engineering experiments, to improve specificity and stability.
- Used to perform rational drug design and design novel proteins.
- Mapping the functions of proteins in metabolic pathways.
- Devise mutagenesis.
The SWISS-MODEL server is a great tool to do homology modelling.
In SWISS-MODEL, the default modelling workflow consists of the following main steps: Input data, Template search, Template selection, Model building and Model quality estimation.
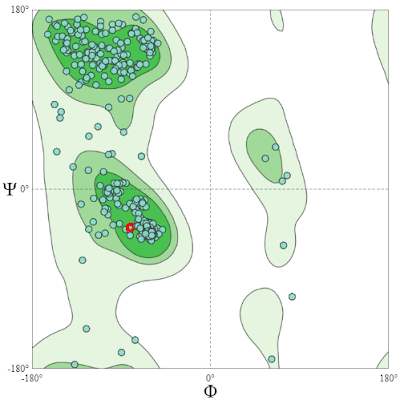
Then the modelled structure can be validated by means of the Ramachandran plot before further analysis.
How to use the SWISS-MODEL server for Homology Modelling?
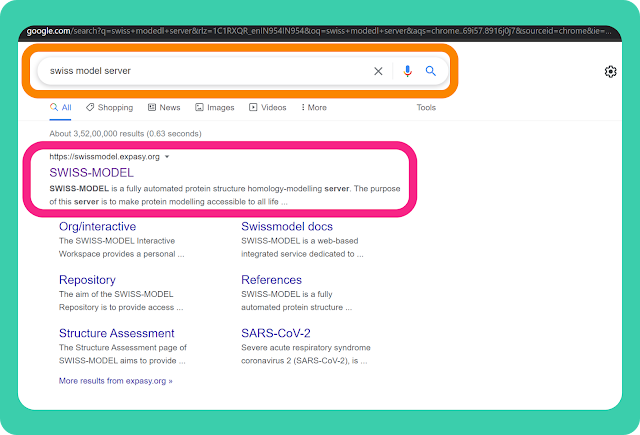
→ SWISS-MODEL server can be accessed through the given URL https://swissmodel.expasy.org/.
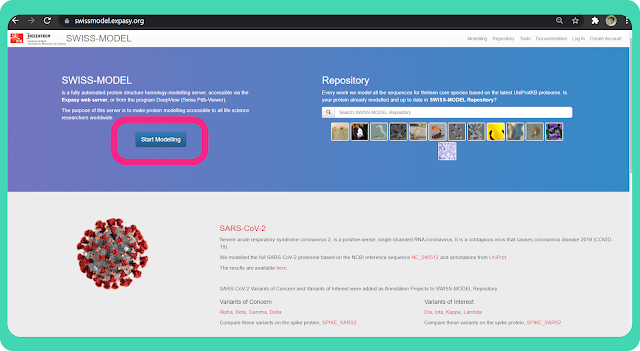
or you can also search "swiss model server" on google. Open the link as marked in the image. It will redirect you to the homepage of "SWISS-MODEL".
→ Click on the "Strat Modelling" option as marked in the image.
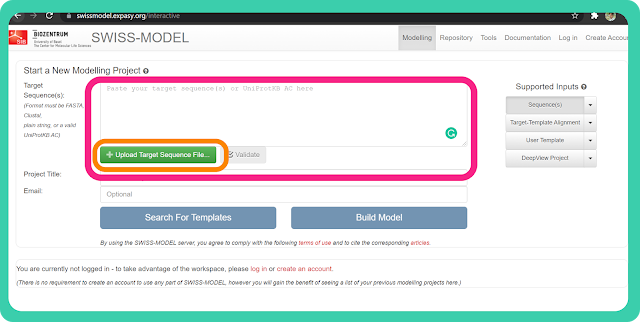
→ It will redirect you to another page (as you can see in the image below).
→ Then you can paste the FASTA sequence in the box or you can also upload the target sequence by clicking on "Upload Target Sequence File".
Here I am using the amino acid sequence of GyrA of Salmonella typhimurium.
UniProt link for GyrA of Salmonella Typhimurium(you can click here and it will redirect you to a UniProt link so that you can get the sequence I have used here).
If you don't know how to retrieve protein sequences from UniProt, I have written another article for that. You can get it through this link (Retrieve Protein sequence and structure) or you can also watch this video https://www.youtube.com/watch?v=ahI0QV_B-sI&t=3s for the same.
For now, you can directly copy the sequence from the box below and use it. But I would suggest you visit Uniprot and try to get the sequence on your own.
>tr|A0A0D6FCL4|A0A0D6FCL4_SALTM DNA gyrase subunit A OS=Salmonella typhimurium OX=90371 GN=gyrA PE=3 SV=1
MSDLAREITPVNIEEELKSSYLDYAMSVIVGRALPDVRDGLKPVHRRVLYAMNVLGNDWN
KAYKKSARVVGDVIGKYHPHGDSAVYDTIVRMAQPFSLRYMLVDGQGNFGSIDGDSAAAM
RYTEIRLAKIAHELMADLEKETVDFVDNYDGTEKIPDVMPTKIPNLLVNGSSGIAVGMAT
NIPPHNLTEVINGCLAYIDNEDISIEGLMEHIPGPDFPTAAIINGRRGIEEAYRTGRGKV
YIRARAEVEADAKTGRETIIVHEIPYQVNKARLIEKIAELVKDKRVEGISALRDESDKDG
MRIVIEVKRDAVGEVVLNNLYSQTQLQVSFGINMVALHHGQPKIMNLKDIISAFVRHRRE
VVTRRTIFELRKARDRAHILEALAIALANIDPIIELIRRAPTPAEAKAALISRPWDLGNV
AAMLERAGDDAARPEWLEPEFGVRDGQYYLTEQQAQAILDLRLQKLTGLEHEKLLDEYKE
LLEQIAELLHILGSADRLMEVIREEMELIRDQFGDERRTEITANSADINIEDLISQEDVV
VTLSHQGYVKYQPLTDYEAQRRGGKGKSAARIKEEDFIDRLLVANTHDTILCFSSRGRLY
WMKVYQLPEASRGARGRPIVNLLPLEANERITAILPVREYEEGVNVFMATASGTVKKTAL
TEFSRPRSAGIIAVNLNDGDELIGVDLTSGSDEVMLFSAAGKVVRFKEDAVRAMGRTATG
VRGIKLAGDDKVVSLIIPRGEGAILTVTQNGYGKRTAADEYPTKSRATQGVISIKVTERN
GSVVGAVQVDDCDQIMMITDAGTLVRTRVSEISVVGRNTQGVILIRTAEDENVVGLQRVA
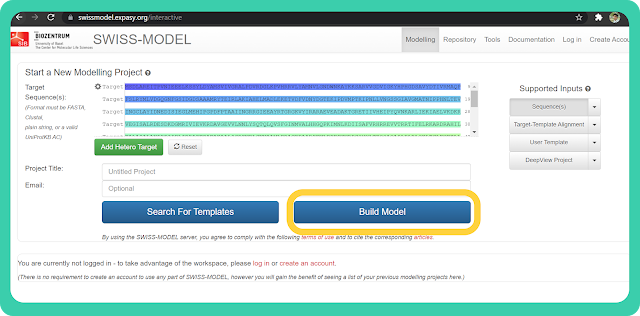
EPVDDEELDAIDGSVAEGDEDIAPEAESDDDVADDADE→ Click on the "Build model" option.
→ It will redirect you to the search result page. Wait for a few minutes. Sometimes it may take more than 10 minutes to show all the results. Make sure that you have a stable internet speed.
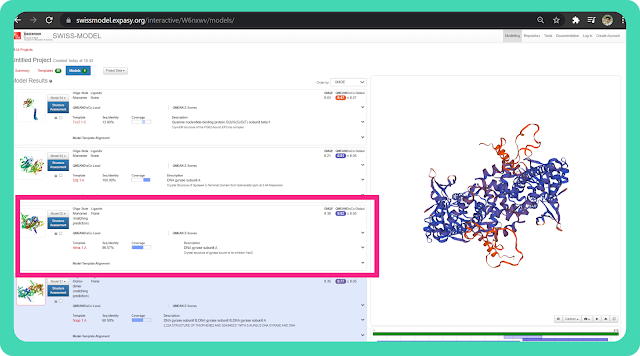
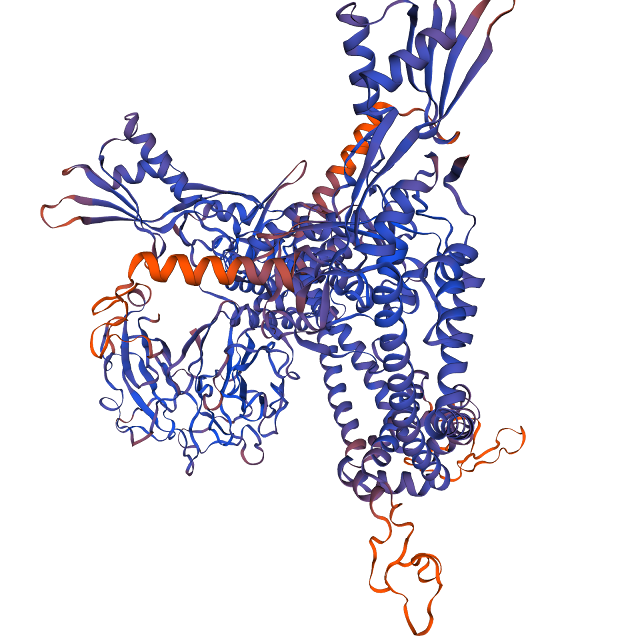
→I am selecting "Model 02" because it has the highest (96.57%) sequence identity as well as a decent amount of coverage.
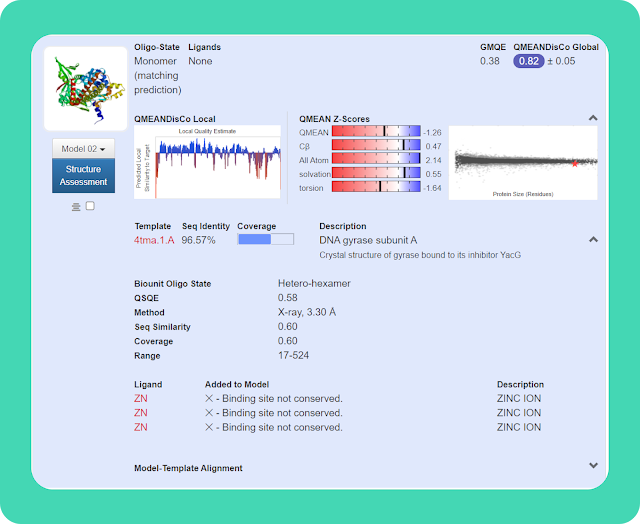
→ You can click on the "Template " option to get more information about the template on which your query sequence get modeled.
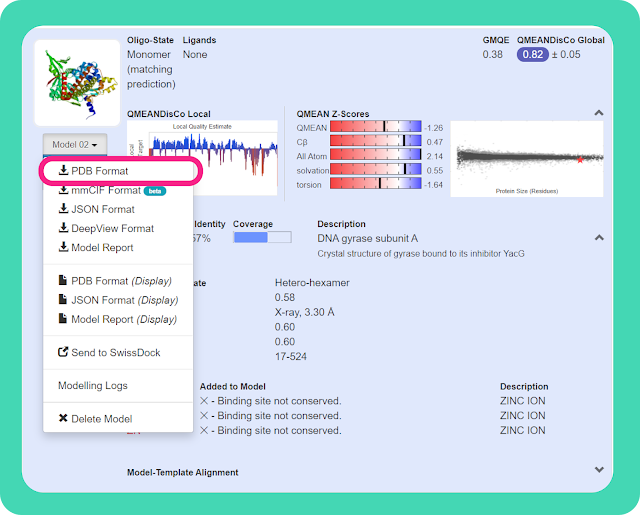
→ Download the modelled structure in the PDB format, which can be later used for validation. You can refer to the image to locate the download option. Remember, you can't open the downloaded PDB file offline without any additional structure visualization software.
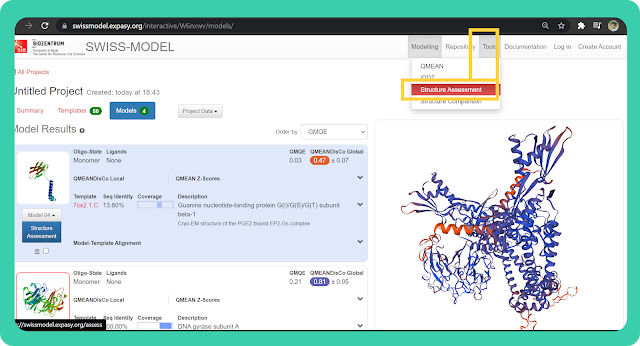
→ To verify the structure, on the basis of the Ramachandran plot, you can go to the "Tools" option in the tab bar. And then select the "Structure Assessment" option. It will redirect you to another page (as shown in the image).
→ Click on "Upload coordinate file" and upload the modelled structure which you have downloaded earlier.
→ Then click on the "Start Assessment" option. You will get all the required data.
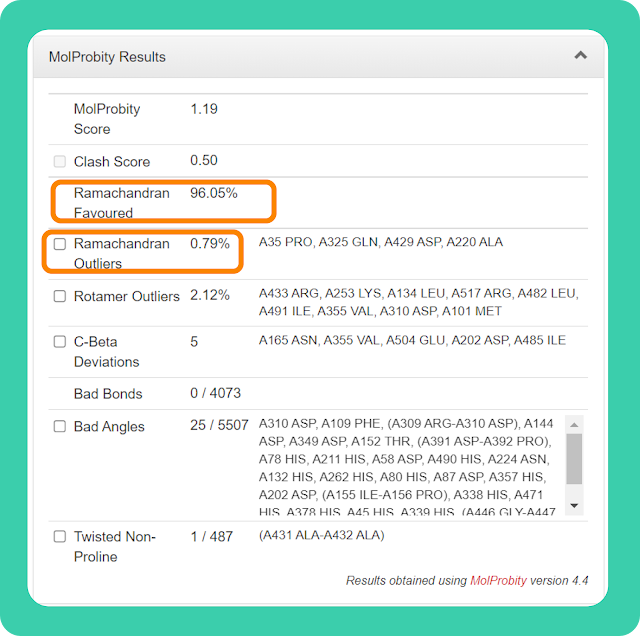
→ As we can see in the image, for my structure Ramachandran favoured value is 96.05% and Ramachandran Outliers value is 0.79% (less than 1%), which is very good. There are other values too but these two values are enough to get an idea about the quality of the structure.
Thank you for reading this article. For any help, you can put a comment below.
by Anant Kumar(Replico)
https://replicoo.blogspot.com/2021/07/retrieving-protein-sequences-and-structure.html
https://replicoo.blogspot.com/2021/07/spyder-python-install.html



Good work
ReplyDeleteGreat job👍👍
ReplyDeleteNice
ReplyDelete